How Can Gene Linkage Be Used To Create Chromosome Maps
Introduction
Since the early 1950s, the development of genetics has been exponential with several landmarks, including determination of DNA as the genetic material in 1944, discovery of the double-helix structure of DNA in 1953, the development of electrophoretic assays of isozymes [1] and a wide range of molecular markers that reveals differences at the DNA level [2]. Each of these milestones had led to a huge wave of progress in genetics. Consequently, our understanding of organismal genetics now extends from phenotypes to molecular levels, which can lead to new or improved screening methods for selecting superior genotypes more efficiently and improve decision-making process in breeding strategies. Genetic mapping (also known as linkage mapping or meiotic mapping) is one of the various applications of molecular markers in any species. It refers to the determination of the relative positions of genes on a DNA molecule (chromosome or plasmid) and of their distance between them. Genetic map indicates the position and relative genetic distances between markers along chromosomes, which is analogous to signs or landmarks along a highway where the genes are "houses" [3] and the first genetic map was published in 1913 by T. H. Morgan and his student, Alfred Sturtevant, who showed the locations of 6 sex-linked genes on a fruit fly chromosome.
The construction of detailed genetic maps with high levels of genome coverage is a first step for some of the applications of molecular markers in plant breeding [4] and serves five purposes: Allow detailed genetic analysis of qualitative and quantitative traits that enable localization of genes or quantitative trait loci [5]. Facilitate the introgression of desirable genes or QTLs through marker-assisted selection; Allow comparative mapping between different species in order to evaluate similarity between genes orders and function in the expression of a phenotype [6,7]. Provide a framework for anchoring with physical maps based on chromosome translocations, DNA sequence or other direct measures [5]. Constitute the first step towards positional or map-based cloning of genes responsible for economically important traits [8]. To be useful for all these purposes, a genetic linkage map has to follow technical and methodological criteria such as simplicity, robustness, transferability, speed and cost effectiveness [9]. The objective/s of the paper was to identify QTL responsible for natural phenotypic variation and assess the principles required for genetic and physical mapping.
Genetic mapping
The early geneticists understood that recombination between genes take place by an exchange of segments between homologous chromosomes in the process now called crossing-over. Each crossing- over is manifested physically as a chiasma, or cross-shaped configuration, between homologous chromosomes; chiasmata are observed in prophase I of meiosis. Each chiasma results from the breaking and rejoining of chromatids during synapsis, with the result that there is an exchange of corresponding segments between them. The theory of crossing-over is that each chiasma results in a new association of genetic markers. Genetic marker is any trait representing genetic differences between individual organisms or species. A genetic map is also called a linkage map or a chromosome map. The linkage of the genes in a chromosome can be represented in the form of a genetic map, which shows the linear order of the genes along the chromosome with the distances between adjacent genes proportional to the frequency of recombination between them. Recombination frequency (also called recombination fraction) between two loci is defined as the ratio of the number of recombined gametes to the total number of gametes produced. Recombination fraction, can be denoted by r, however, has a domain of 0 ≤ r ≤ 0.5, with r= 0 indicating perfect linkage and r = 0.5 meaning complete independence of the two loci [10]. For the successful construction genetic map there are principles we should follow and there are requirements for mapping that are mapping populations, selection of molecular markers for mapping, polymorphism screening and genotyping of the mapping population and linkage analysis. In this section we will try to see each point as much as possible Figure 1.
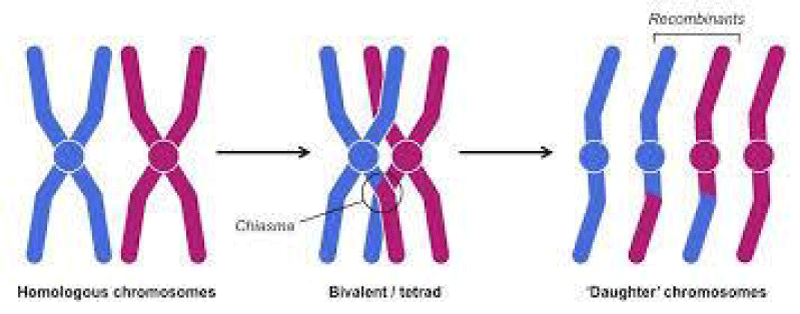
Figure 1:Crossing-over of homologus chromosomes during prophase ׀ of meiosis.
Source: Chiasma (genetics) - Wikipedia en.wikipedia.org.
Principles of genetic mapping
Genetic mapping is based on the principle that genes (markers or loci) segregate via chromosome recombination during meiosis (i.e. sexual reproduction), thus allowing their analysis in the progeny [11]. During meiosis, chromosomes assort randomly into gametes, such that the segregation of alleles of one gene is independent of alleles of another gene. This is stated in Mendel's second law and is known as the law of independent assortment. The law of independent assortment always holds true for genes that are located on different chromosomes, but it does not for genes that are on the same chromosome. Genes that are closer together or tightly-linked will be transmitted together from parent to progeny more frequently than those genes located far apart. At the beginning of meiosis, a homologous chromosome pair may intertwine form so called chiasma, plural chiasmata and exchange sections of chromosome [12]. Such process or set of processes is called recombination (also called cross-over or strand exchange) by which DNA molecules interact with one another to bring a rearrangement of the genetic information in an organism. The pair then breaks apart to form gametes with new combination of genes that differs from either of the parents.
The observed recombination fraction between two loci is an estimate of one-half the number of chiasmata or crossover events between two loci because crossing over occurs at the four-strand stage and for single crossover events, only two of the four strands participate in the recombination. Two non-sister chromatids participate in the cross-over and the other two chromatids do not exchange chromosome segments. Such process produces two types of gametes: If crossing over does not occur, the products are parental gametes and if crossing over occurs, the products are recombinant gametes. The allelic composition of parental and recombinant gametes depends upon whether the original cross involved genes in coupling or repulsion phase. In diploid species, the most prevalent gametes in a coupling phase will be those with two dominant alleles or those with two recessive alleles. For repulsion phase crosses, gametes containing one dominant and one recessive allele will be most abundant. Genes located on different chromosomes assort independently (unlinked) and have a recombination frequency of 50%, linked genes have a recombination frequency that is less than 50% [13]. The chance of a crossover producing recombination between genes is directly related to the distance between two genes the lower the frequency of recombination between two markers, the closer they are situated on a chromosome (conversely, the higher the frequency of recombination between two markers, the further away they are situated on a chromosome).
Requirements for genetic mapping
Mapping population: The first step in producing a mapping population is selecting two genetically divergent parents, which show clear genetic differences for one or more traits of interest. They should be genetically divergent enough to exhibit sufficient polymorphism and at the same time they should not be too genetically distant so as to: a) Cause sterility of the progenies and/or b) Show very high levels of segregation distortion during linkage analysis. In self-pollinating species, mapping populations originate from parents that are both highly homozygous (inbred). In cross pollinating (outcrossing) species, the situation is more complicated since most of these species do not tolerate inbreeding. Selection of populations is critical to successful linkage mapping. F2 populations are developed by selfing F1 hybrids derived by crossing the two parents while BC population is produced by crossing F1 back into one of the parents (the recipient or recurrent parent). Recombinant Inbred Lines (RILs) are developed by single-seed selections from individual plants of an F2 population; such selections continue for six to eight generations. If backcross selection is repeated at least for six generations, more than 99% of the genome from BC6 and above will be derived from recurrent parent [14]. Selfing of selected individuals from BC7F1 will produce BC7F2 lines that are homozygous for the target gene, which is said to be nearly isogenic with the recipient parent (NILs). Nearly isogenic lines are frequently generated by plant breeders as they transfer major genes between varieties by backcross breeding [4] Figures 2,3.
A Double Haploid (DH) population is produced by doubling the gametes of F1 or F2 population. Plants will be regenerated using tissue culture techniques after induction of chromosome doubling from pollen grains or haploid embryos resulting from species crosses. RILs, NILs and DHs are permanent populations because they are homozygous or 'true-breeding' lines that can be multiplied and reproduced without genetic change occurring. Seeds from RILs, NILs and DHs can be transferred between different laboratories for linkage mapping to ensure that all collaborators examine identical material [11,15,16]. Double Haploid (DH) populations are quicker to generate than RILs and NILs but the production of DHs is only possible for species with a well-established protocol for haploid tissue culture. The time required for developing RILs and NIL populations is a major constraint in mapping studies. In practice, the population size used in preliminary genetic mapping studies varied from 50 to 250 individuals [17], but a larger population size is needed for high resolution fine mapping.
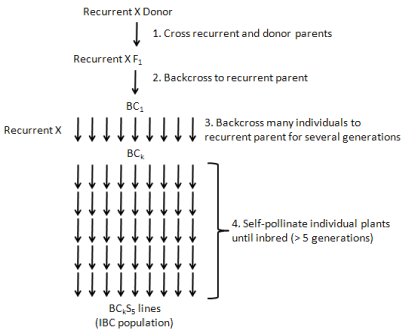
Figure 2:Schematic illustrating the development of an inbred backcross (IBC) population
Source: Matthew Robbins, the Ohio State University.
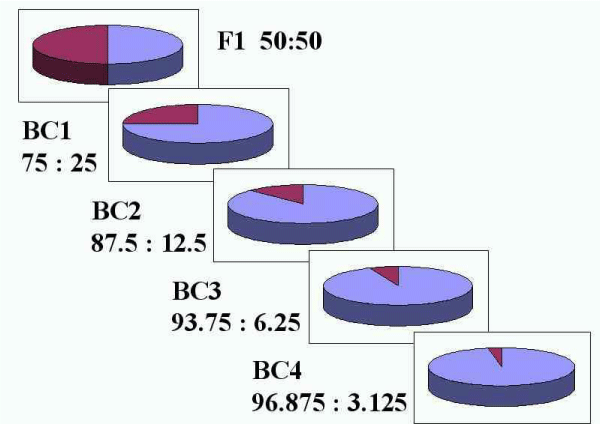
Figure 3:The contribution of the donor parent genome is reduced by half with each generation of backcrossing.
Source: David Francis, the Ohio State University.
Selection of molecular markers for mapping
In traditional plant breeding, genetic diversity was usually diagnosed through observational selection. But now, with the development of molecular biology, this work is determined at molecular level based on DNA changes and their effects on the phenotype. Once DNA was extracted from plant, changes in the samples are determined using PCR or hybridization and subsequent agarose or acrylamide gel electrophoresis to recognize different molecules based on their size, chemical composition or charges [18,19]. Genetic markers are biological compounds which can be determined by allelic variations and can be used as experimental probes or labels to track an individual, tissue, cell, nucleus, chromosomes or genes. In classical genetics, genetic polymorphism represents allele diversity. While in modern genetics, genetic polymorphism is the relative difference in genetic locus of the genome. Genetic markers can be used to facilitate the study of heredity and variation [20]. DNA markers involve several sets of markers and divided into two main categories: PCR-based molecular markers and hybridization-based molecular markers. Restriction Fragment Length Polymorphism (RFLP) is a hybridization-based molecular marker while the other markers like microsatellites or Simple Sequence Repeats (SSRs), Expressed Sequence Tags (ESTs), Cleaved Amplified Polymorphic Sequence (CAPS), Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphisms (AFLPs), Inter Simple Sequence Repeat (ISSR), diversity arrays technology (DArT), and Single Nucleotide Polymorphism (SNP) are all PCR-based [4] and have been used for map construction in several plants. The first large scale efforts to produce genetic maps were performed mainly using RFLP markers, the best known genetic markers at the time [21]. The major strength of RFLP markers includes codominant inheritance, good transferability between laboratories, locus-specificity that allows synteny (conserved order of genes between related organisms) studies, and high reproducibility. There are, however, several limitations for RFLP analysis: that it requires high quantity and quality of DNA; depends on the development of specific probe libraries for the species; the technique is not amenable for automation; the level of polymorphism is low; it is time consuming and laborious; it usually requires radioactively labeled probes. With the development of PCR-based markers, the strategy in linkage mapping dramatically shifts to new type of marker and currently, microsatellite markers remain a standard for linkage map construction. The advantages of SSRs are well documented [22,23] and these include: high information content, co-dominant inheritance, reproducibility, locus specificity, highly transferability, and ease for automation for high throughput screening. However, the high development cost and effort required to obtain working SSR primers for a given species has restricted their use to only a few of the agriculturally important crops [24].
Polymorphism screening and genotyping of the mapping population
The third step in the construction of a linkage map is to identify sufficient number of markers that reveal differences between parents (i.e. polymorphic markers). It is critical that sufficient polymorphism exists between parents in order to construct a linkage map. In general, cross pollinating species possess higher levels of polymorphism compared to inbreeding species. Once sufficient numbers of polymorphic markers have been identified between parents, they must be used to genotype the entire mapping population. The progenies will inherit a marker from either of the parents (homozygous dominant or homozygous recessive) or both parents (heterozygous) but the scoring method is different between codominant and dominant markers, as the latter is not able to show differences between dominant homozygous and heterozygous individuals.
A genetic map is as good as the data that were used to construct it. Researchers construct a linkage map assuming no errors present in the data and then look for improbable genotypes, such as those originating from double recombination. Therefore, the data must be critically checked for all possible errors, such as typographical error, missing data, genotype coding error, order of genotypes along all loci, etc. It is absolutely essential that the order of the individuals is identical over all loci in the data file. As discussed by several authors [25,26], a low frequency of typing errors can have a substantial impact on the order and length of a linkage map. The most likely effect of a typing error is to introduce a double recombination, so that an individual's genotype at three neighboring loci might change from a true genotype of ABA to ABB. This is increasingly the case as the marker density increases and the proportion of true recombinations between neighboring markers falls. Individuals with too much missing data will contribute very little information in the map calculations; in fact they might even cause problems [27]. The presence of missing values in the marker data means that information about the number of true recombination that has taken place along the chromosome is lost. Hackett and Broadfoot [26] performed a simulation study to investigate the effects of typing errors and missing values on the construction of linkage maps and concluded that missing values had less effect than typing errors, but they reduced the number of correctly ordered markers and produced shorter map lengths for more widely separated markers.
Linkage analyses and map construction
Linkage analyses and mapping are computerized. Several computer packages are presently available for genetic linkage mapping but the most widely used are Join Map [28], Map Maker/Exp [29], Linkage [30] and Map Manager QTX [31]. Join Map is a commercial program while all others are freely available from the internet. The basic principles in map construction are basically the same for the different statistical programs, and the major steps in linkage analyses are described using Join Map as an example.
A) Test for segregation distortion
For each segregating marker, a chi-square analysis needs to be performed to test for deviation from the expected segregation ratio for the mapping population (1:1 for both dominant and codominant markers in BC, RIL, DH and NIL; 1:2:1 for codominant markers in F2; 3:1 for dominant markers in F2). A deviation of the observed genotypic frequencies from the expected in a given genotypic class within a segregating population is called segregation distortion [32-34]. Segregation distortion can occur due to different reasons: statistical bias, genotyping and scoring errors [35] and biological reasons like chromosome loss, competition among gametes for preferential fertilization, gametocidal or pollen-killer genes (abortion of male or female gametes), incompatibility of genes, chromosome arrangements or non-homologous pairing [34,36,37]. Segregation distortion is a problem often encountered in mapping studies [38,39]. It has been shown that the analysis of linkage may be influenced by deviations of single locus segregation ratios from expected frequencies, and several authors have discussed methods to test for linkage or to estimate recombination frequencies between genes showing segregation distortion [40].
The effects of inclusion of loci with significant segregation distortion in the final linkage map seem contradictory. According to Hackett and Broadfoot [26], segregation distortion had very little effect both on marker order and map length but others have reported reduction in map length due to the presence of loci with significant segregation distortion. Segregation distortion is a normal phenomenon in wide crosses, and one should be careful in removing loci with segregation distortion from further calculation. It is better to study these loci after calculating the map, as markers distorted towards the same direction clustered in a small chromosome region [41]. In plants, the percentage, degree, origin and genetic effects of segregation distortion vary significantly with species, population types, crosses and marker types. DH and RIL populations usually have high segregation distortion while BC populations usually have relatively fewer segregation distortions. For DH, non-Mendelian segregation may arise due to the various reasons mentioned above plus selection associated with the in vitro DH production process and fixation of recessive lethal genes into homozygous. In view of molecular marker types, Lorieux, et al. [40] indicated that the estimation of recombination fractions in codominant markers is less affected by segregation distortion than that of dominant markers. In most cases, however, the scale and extent of segregation distortion for codominant versus dominant markers varied significantly among different data.
B) Establishing linkage groups
Markers are assigned to linkage groups using the odds ratios, which refers to the ratio of the probability that two loci are linked with a given recombination value over a probability that the two are not linked. This ratio is called a Logarithm of Odds (LOD) value or LOD score [42,43]. The critical LOD scores used to establish linkage groups and calculate map distances are called 'linklod' and 'maplod', respectively [42,44]. Marker pairs with a recombination LOD score above a critical 'linklod' are considered to be linked whereas those with a LOD score less than 'linklod' are considered unlinked. Several researchers used a 'linklod' value of 3 as the minimum threshold value in order to decide whether or not loci were linked. A LOD value of 3 between two markers indicates that linkage is 1000 times more likely than no linkage (Stam, 1993a). Experience with modern data sets with many markers, especially those species with large numbers of chromosomes, shows that even using a 'linklod' of > 6 may lead to false positive linkage [42,45].
Higher critical LOD values will result in more number of fragmented linkage groups, each with smaller number of markers while small LOD values will tend to create few linkage groups with large number of markers per group. Two markers are placed in distinct linkage groups if they are not linked to any member of the other group. At any stage in the calculation, there is a group of markers which have been assigned to a linkage group and a group of free markers which have not yet been assigned. Various options (e.g. changing the parameters of analyses, excluding loci or individuals, generation of additional marker data for linkage groups with few number of markers) can be tested until researchers establish satisfactory linkage groups. Ideally one would like to arrive at a number of linkage groups that is the same as the haploid chromosome numbers of the species under study. In practice, determining number of linkage groups is usually not a straightforward task because; i. Loci on different chromosomes may appear to be linked by chance (spurious linkage). ii. Two or more linkage groups can be obtained for each chromosome, which results to the total number of linkage groups much higher than the haploid chromosome numbers.
If a linkage group consists of loci from different chromosomes, this often leads to many suspect linkages and to a poor goodness-of-fit of the resulting map. Therefore, it is important to critically check every marker in each linkage group before proceeding to the next step. Furthermore, the number of linkage groups, especially in polyploidy species [45], can be higher than the number of haploid chromosomes if the molecular markers are not well distributed across all chromosomes and do not sufficiently covers the genome. The next step, after establishing linkage groups, is to assign them to a chromosome based on previous available information for anchoring markers (markers with known chromosomal location) and/or using aneuploidy lines. Of course, the former is not possible if one starts from scratch. If there are no previously mapped anchoring markers, one needs to determine the chromosomal location of selected markers in a linkage group using aneuploid lines, such as nullisomics, monosomics, and chromosome deletion stocks [46]. Aneuploid is the condition in which the chromosome number of the cells of an individual is not an exact multiple of the typical haploid set for that species. If a marker is missing from a certain chromosome (piece) in aneuploid lines, it is strong evidence for its physical location. Aneuploids are very useful for assigning linkage groups of genetic markers to both physical chromosomes as well as for merging two or more linkage groups that belong to the same chromosome. However, one should remember that a single probe or primer-pair may produce multiple loci, especially in polyploidy, and the different loci may map to different chromosomes.
C) Determining map distance and locus order
For calculating map distances and determining locus order, the researchers need to specify several parameters, including a recombination threshold value, minimum 'maplod', jump threshold value, and mapping function (m.f.). Only information for marker pairs with a LOD score above, 'maplod' is used in the calculation of map distances. The choice of 'maplod' values is arbitrary and it can be as low as 0.01 to as high as 3.0. If the value of 'maplod' equal to 0.01, the program uses even very weak linkage information (usually corresponding to recombination values slightly less than 50%). If one is dealing with large linkage groups (i.e., over 50 markers per linkage group), various marker pairs will show insignificant linkage, if at all. In such cases, the 'maplod' value should be set between 0.5 and 1.0 to ensure that no information is used which comes from distant markers [43]. The mapping procedure is basically a process of building a map by adding loci one by one, starting from the most informative pair of loci (loci pair with most linkage information). If the order of sets of (at least three) markers is known in advance, this information can be provided to the program as a "fixed order" [27,43,47].
Relationship between genetic and physical maps
The order of markers in a high density genetic map remains the same as that of the physical map. However, there is no direct linear relationship between units of genetic distances in centimorgan (cM) and physical distances in kilo base pairs (kb). For chromosome 4 of Arabidopsis, for example, the kilo base-pair to centimorgan ratio varied from 30 to 550 kb per cM [48]. In rice 1 cM on average equals to 258.5 kb [49], but this figure actually varies from 120 to 1000 kb per cM. In wheat, the variation is even more extreme, with 1 cM equivalent to 118 to 22,000 kb. Therefore, genetically close markers may actually be far apart in terms of base pairs (or vice versa) due to differences in the frequency of recombination along the length of a chromosome. The nonlinear relationship between genetic and physical map distances can hinder the ability of geneticists to identify genes by map-based techniques. Understanding the rules that govern the distribution of recombination events will be of great value to researchers who aim at identifying genes on the basis of their position in a genetic map. Meiotic recombination occurs preferentially at defined sites, termed hot spots, along chromosomes of various eukaryotic organisms [50].
In all eukaryotic organisms that have been analyzed in detail, regions of high (hot spots) and low (cold spots) recombination have been reported [50,51]. In hexaploid wheat, more than 85% of the genes are present in gene-rich regions, physically spanning only 5-10% of the chromosomal region [52]. The gene-rich regions are interspersed with blocks of repetitive DNA sequences visualized as regions of low gene density. These gene-rich regions undergo recombination much more frequently than do gene-poor regions. Kilo base-pair per cM estimates ranged from 118 kb for gene-rich regions to 22,000 kb for gene-poor regions [52]. Physical distribution of recombination events is nonrandom in other plant species as well [53]. The genetic- and physical-map orders of markers are not without errors. Inaccuracies in genetic maps can result from genotyping errors, as well as from the use of a limited number of informative meiosis to generate maps. Errors in the order of markers on physical maps can be due to problems with assembly or to incorrect identification of marker positions. Even when the order of markers is known to be without error, accurate estimates of recombination fractions will play an important role in linkage studies [54].
Prospects of genetic mapping
The development of genetic maps based on markers that are simple to generate, highly reproducible, codominant, and specific for known linkage groups are highly desirable for their application in breeding. The transferability of maps constructed using AFLPs, RAPDs, and ISSRs is limited between populations and pedigrees within a species [55], because each marker is primarily defined by its length (i.e., sequence information may be limited). Moreover, the same size band amplified across populations/species does not necessarily mean that bands possess the same sequence, unless proven by hybridization studies [56]. In contrast, the development of high density maps that incorporate EST-derived RFLP, SSR and SNP markers will provide researchers with a greater arsenal of tools for identifying genes or QTLs associated with economically important traits. Furthermore, such EST-based markers mapped in one population can be used as probes and primers for characterizing other populations within the same species.
Genetic maps with good genome coverage and confidence in locus order requires not only large numbers of DNA markers, but also the analyses of large numbers of individuals. The requirement for a large number of markers or mapping populations to reduce the linkage group number to haploid chromosome numbers and increase map accuracy has been emphasized in mapping studies [57-59]. The methods for detection and analysis of widely-used markers are automated and much faster than some years ago. One example of an improvement in the efficiency of marker analysis is multiplex PCR, which enables multiple marker loci to be tested simultaneously. PCR products up to 9 different primer pairs with non-overlapping ranges of allele sizes can be multiplexed and run on high throughput DNA sequencing machines. New types of high-throughput marker systems, such as SNPs, should play an important role in the construction of high density maps, provided that these methods are not too expensive.
Due to the abundance of SNPs and development of sophisticated high throughput SNP detection systems, it is expected that SNP markers will have a great influence on future mapping studies [60]. Comparison of sequences of the two rice cultivars 'Nipponbare' (japonica) and 'Kasalath' (indica) revealed a total of 80,127 polymorphic SNPs and 18,828 potentially polymorphic SSRs. This suggests the high potential of SNP and SSR markers for other species as well for the construction of highly saturated genetic maps. Genetic maps based on DNA markers are available for several economically important plants, including Arabidopsis, maize, rice, wheat, barley, tomato, potato, sunflower, pea, bean, rye, millet, cotton, soybean, sorghum, cowpea, tobacco, turnip rape, cauliflower, sunflower, alfalfa, carrot, sugarcane, sugar beet, coffee, and grape. In model species (such as Arabidopsis and rice) and other extensively studied species, one can find several genetic maps developed by different researchers using different mapping populations, sample size, marker systems, and statistical programs. However, it is not always possible to get the same map length and marker order in these different genetic maps. Such type of problem can be solved with the development of a wide range of high throughput techniques for physical mapping of chromosomes.
Summary and Conclusion
Genetic maps are important genomic resources for genomic and genetic studies, which were widely used for genome assembly, functional gene mapping and comparative genome analysis. Gene mapping is the sequential allocation of loci to a relative position on a chromosome. Genetic maps are species-specific and comprised of genomic markers and the genetic distance between each marker. The genetic base of crop production can be preserved and widen by an integration of biotechnology tools in conventional breeding. Similarly targeting specific genotypes to particular cropping systems may be facilitated by understanding specific gene-by-environment interaction(s) with the aid of molecular research. Agricultural genetics is the applied study of the effects of genetic variation and selection used to propagate valuable heritable trait combinations in crop plants and farm animals. The great advantage of genetic mapping is that it can identify the relative position of genes based solely on their phenotypic effect. Genetic mapping is a way to identify exactly which chromosome has which gene and exactly pinpointing where that gene lies on that particular chromosome.
Genetic mapping offers evidence that a disease transmitted from parent to child is linked to one or more genes and provides clues about which chromosome contains the gene and precisely where the gene lies on that chromosome. Genes that are located on the same chromosome are called linked genes. Detection of the marker can be direct by RNA sequencing, or indirect using allozymes. Some of the methods used to study the genome or phylogenetic are RFLP, AFLP, RAPD, SSR. They can be used to create genetic maps of whatever organism is being studied. Genetic and physical maps illustrate the arrangement of genes and DNA markers on a chromosome. The relative distances between positions on a genetic map are calculated using recombination frequencies, whereas a physical map is based on the actual number of nucleotide pairs between loci. Linkage mapping, a conventional mapping method, depends upon genetic recombination during the construction of mapping populations. The phenotypic variation of many complex traits of agricultural or evolutionary importance is influenced by multiple Quantitative Trait Loci (QTLs), their interaction, the environment, and the interaction between QTL and environment.
Generally, genome mapping is a widely-applicable approach to scanning the genetic information of an organism for genes that are responsible for a specific trait. With the rapid increase in the world population, the demand for food is also increasing and there is a need to develop high-yielding varieties with more resistance to biotic and abiotic stress. There is a need to precisely correlate genotype with phenotype. Therefore, the order and relative distance of genetic features that are associated with genetic variation or polymorphisms can be determined by genetic mapping to detect the desirable traits for boosting productivity. Genetic maps constructed using molecular markers can also be used to locate major genes which can then also be used as genetic markers. In classical genetics, genetic polymorphism represents allele diversity, while in modern genetics, genetic polymorphism is the relative difference in genetic locus of the genome.
How Can Gene Linkage Be Used To Create Chromosome Maps
Source: https://www.peertechzpublications.com/articles/OJPS-6-128.php
Posted by: englesdoony1936.blogspot.com

0 Response to "How Can Gene Linkage Be Used To Create Chromosome Maps"
Post a Comment